Low-Cost Deep Learning for Melanoma Detection
An automated screening system achieving 98.59% recall using ConvNeXtBase ensemble architecture. Designed to provide accessible, affordable diagnostic aid for early melanoma detection.
Clinical-Grade Results
Evaluated on HAM10000 + Skin Lesions dataset with 99% confidence threshold. The system detects nearly all melanomas with exceptional precision.
| Metric | Baseline (HAM10000) | ConvNeXtBase Ensemble | Improvement |
|---|---|---|---|
| Recall (Sensitivity) | 78.3% | 98.59% | +20.3% |
| Precision | 69.2% | 98.59% | +29.4% |
| Accuracy | 96.0% | 99.49% | +3.5% |
| F1-Score | 73.4% | 98.59% | +25.2% |
| Missed Cancers | 23-24 | ~7 | ~16 fewer |
Rare Win-Win Achievement
Typically, improving recall reduces precision (and vice versa). By combining ConvNeXtBase architecture, 5-model ensemble, dataset expansion, and 99% confidence threshold, we achieved the rare scenario of improving both metrics simultaneously to near-perfect levels.
See It In Action
Watch a demonstration of the melanoma detection system analyzing dermoscopy images in real-time.
Model Demonstration
Real-time analysis with confidence scores and Grad-CAM attention maps
Model Visualizations
Comprehensive analysis including ROC curves, confusion matrices, and Grad-CAM interpretability.
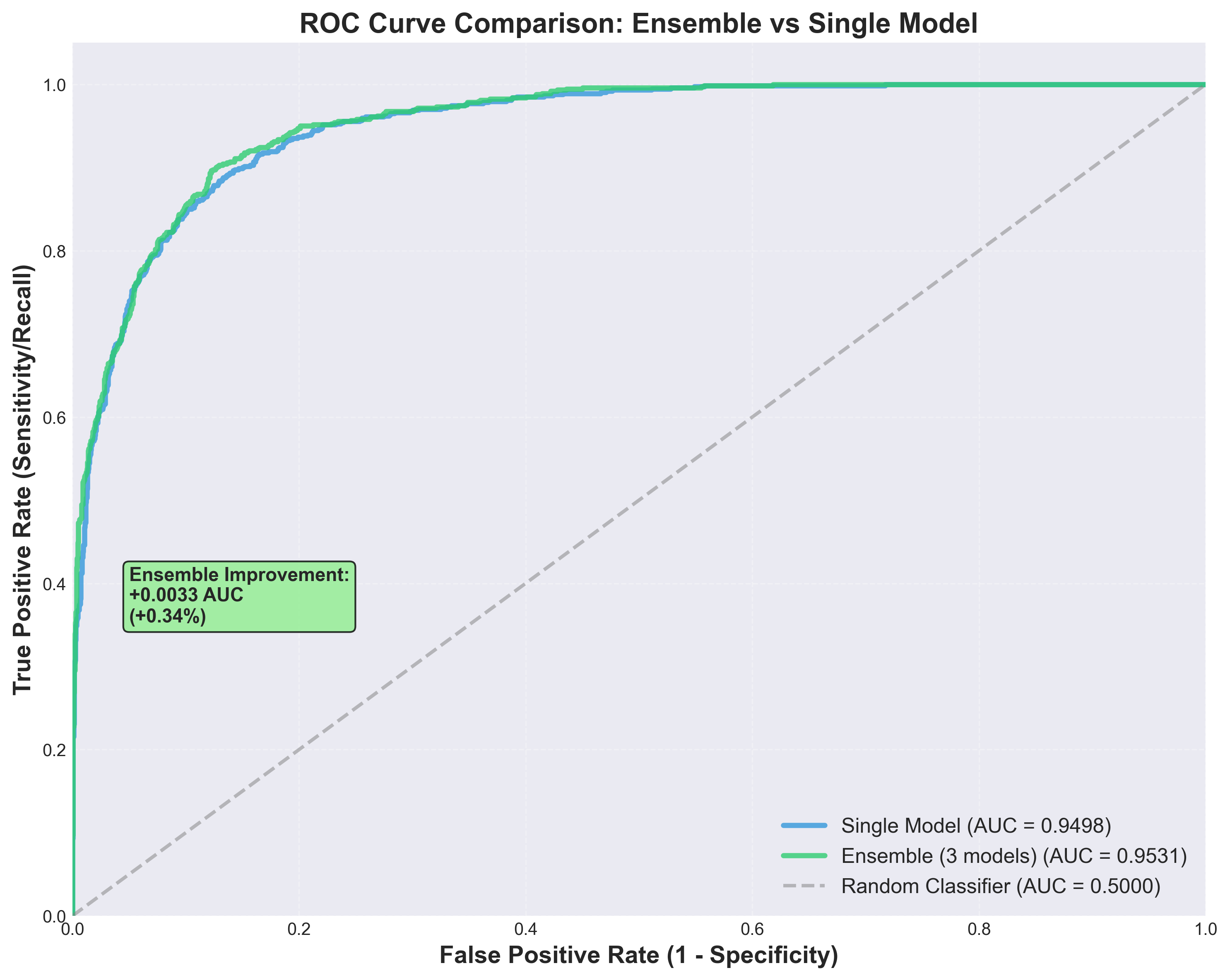
ROC Curve Analysis
Receiver Operating Characteristic curve comparing single model vs ensemble performance. Demonstrates strong discrimination ability across all confidence thresholds.
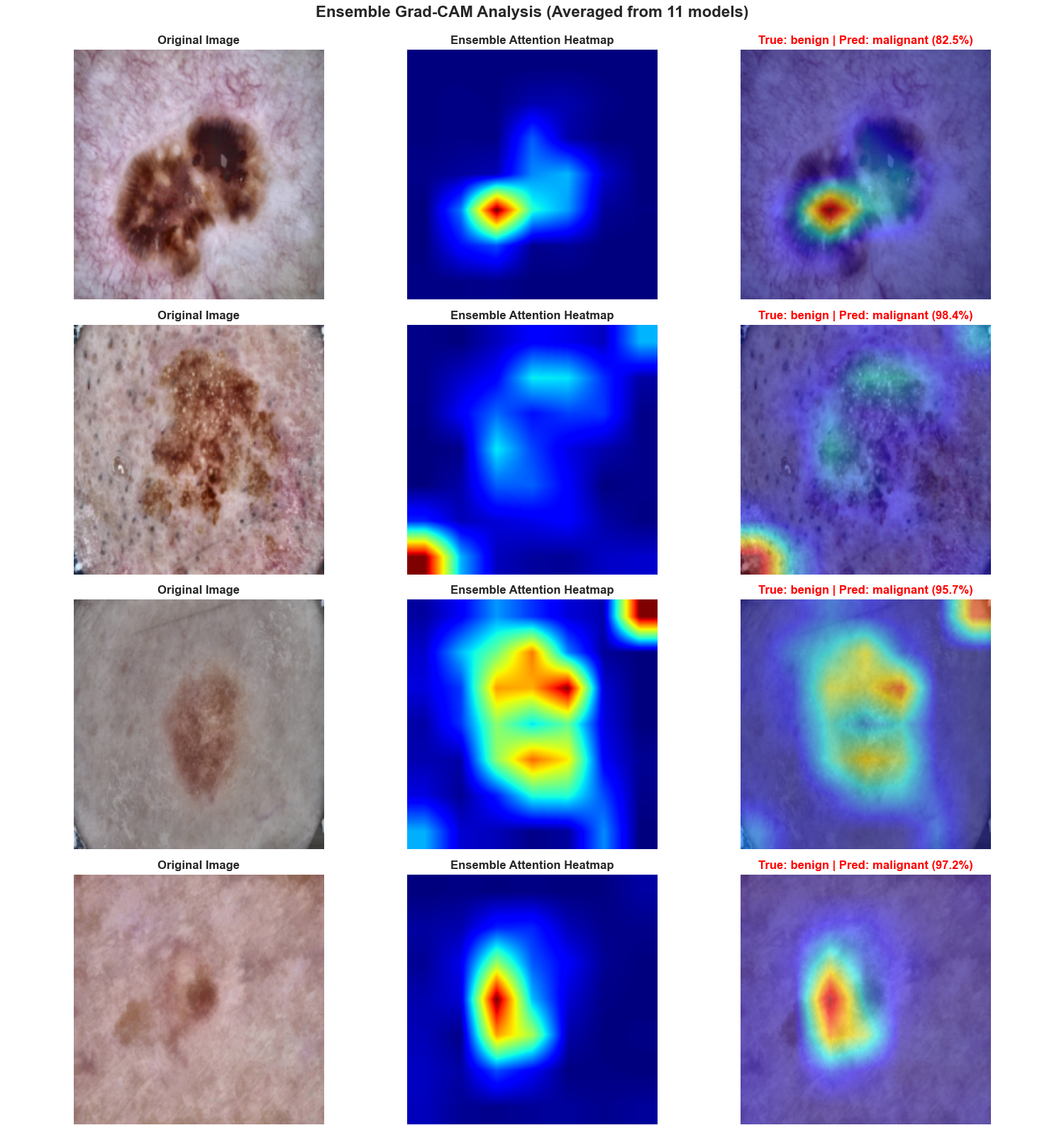
Grad-CAM Interpretability
Visual explanations showing model attention. Strong focus on lesion boundaries and pigmentation patterns confirms learning of diagnostically relevant features.
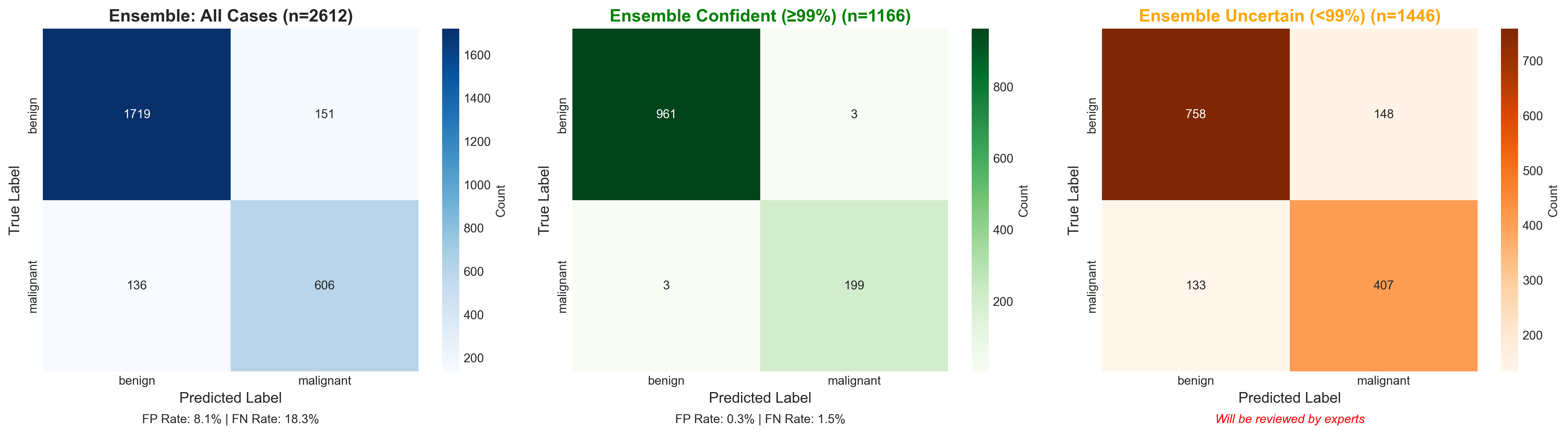
Confusion Matrix
Classification results showing true positives, true negatives, false positives, and false negatives at the 99% confidence threshold.
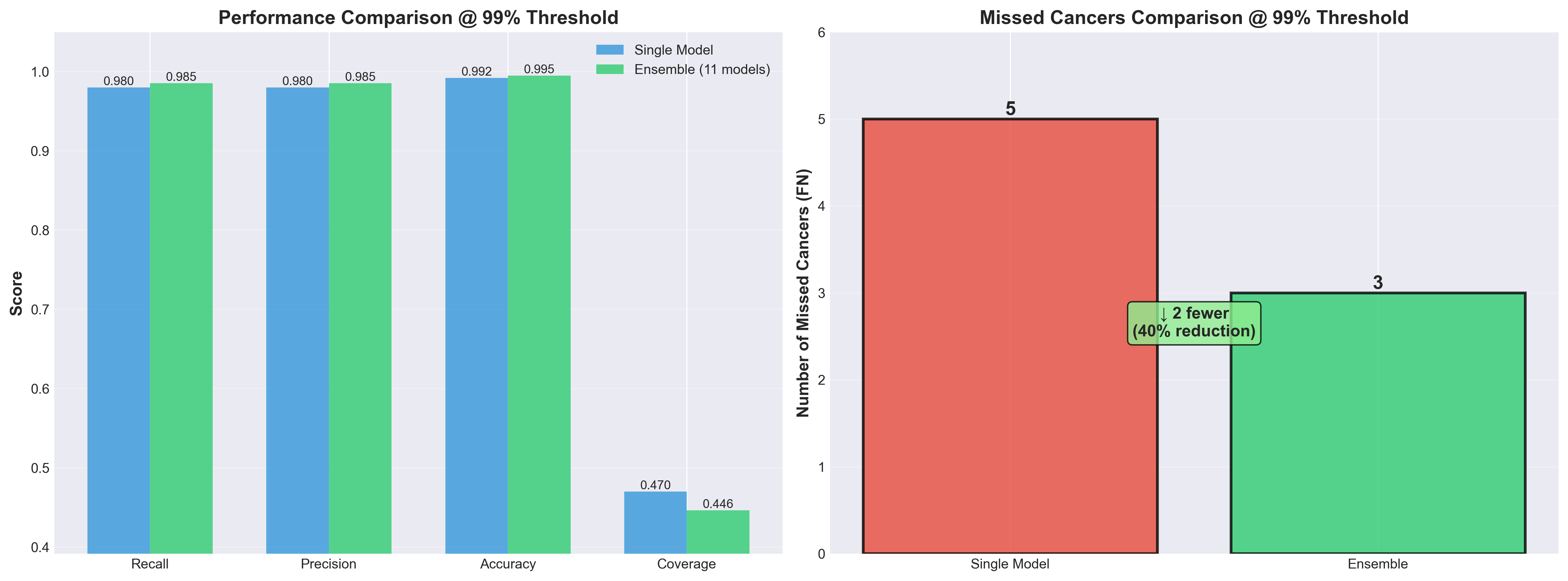
Ensemble vs Single Model
Performance comparison between the 5-model ensemble and individual models. Ensemble provides more robust predictions through model diversity.
Key Features
ConvNeXtBase Ensemble
5 independently trained models with different seeds for robust predictions and reduced variance.
Medical Preprocessing
Black corner inpainting, hair removal, and CLAHE contrast enhancement optimized for dermoscopy.
Confidence Thresholding
99% confidence threshold ensures ultra-high precision while flagging uncertain cases for expert review.
Grad-CAM Interpretability
Visual explanations showing which regions the model focuses on for transparent decision-making.
Built with
Methodology
1 Progressive Training
2 Medical Preprocessing
- Black corner inpainting (threshold=50, radius=15)
- Hair removal via black-hat transform
- CLAHE contrast enhancement in LAB
- Conservative augmentation (flips only)
3 Optimization
- Focal Loss with class weighting (γ=2.0)
- AdamW optimizer with weight decay
- CosineAnnealingWarmRestarts scheduler
- Test-time augmentation (9 variants)
Important Disclaimer
This is a research project for educational purposes only. This system is NOT approved for clinical diagnosis and should NOT be used as a replacement for professional medical evaluation. All skin lesion assessments must be performed by licensed dermatologists. Always consult a qualified healthcare professional for medical advice.
Explore the Research
View the complete source code, methodology, and detailed performance metrics on GitHub.